I came across an interesting report on people who are "face blind" (Prosopagnosia) on CBS 60 minutes show today and thought to share it.
Face Blindness: When everyone is a stranger - CBS News
http://www.cbsnews.com/8301-18560_162-57399118/face-blindness-when-everyone-is-a-stranger/
Do you have trouble recognizing faces? Take a test - 60 Minutes Overtime - CBS News http://www.cbsnews.com/8301-504803_162-57399227-10391709/do-you-have-trouble-recognizing-faces-take-a-test/
The researchers at Harvard University has two tests to check face recognition ability . Here are the online tests to access how good you are in recognizing faces. Have a look at them and take it if you have 20-30 minutes to spare.
http://www.faceblind.org/facetests/index.php
Some other brain tests:
BBC - Science & Nature - Human Body and Mind - Face Memory Test http://www.bbc.co.uk/science/humanbody/sleep/tmt/
It's amazing to think about the intricacies and mysteries of brain function. I'm looking forward to the science advancements in this field.
InsilicoScientist blogs: Digital world of Drug design and Discovery
Some of the interesting aspects to ponder relevant to Computer Aided Drug Design(CADD)
Sunday, August 05, 2012
Saturday, July 23, 2011
Spicy hot long pepper could kill cancer cells
Natural products have played a significant role in fighting cancer. Taxol and Vinca alkaloids are examples of currently approved natural product drug for the treatment of cancer. Many more natural products like curcumine, the active ingredient of South Asian spice turmeric, have been shown to possess anticancer properties. The latest addition to the list is a constituent of long pepper. Piperlongumine (PL), a constituent of the fruit of a pepper plant found in southern India and southeast Asia (Piper longum; തിപ്പലി, പിപ്പലി (tippali, pippali) in Malayalam) has been shown to possess anti-cancer properties. It selectively targets and kills cancer cells but leaving normal cells unharmed.
As a computational medicinal chemist, I was curious about the potential molecular targets of Piperlongumine. From the first look at the chemical structure, it appeared to have some similarity (trimethoxyphenyl group) to Colchicine, a tubuline targeting small molecule.
However the authors found that Piperlongumine acts differently in inducing ROS than the tubuline binding agent Taxol. The compound is reported to target the ROS stress-response pathway. A search to the PubChem chemical biology repository identified shows 22 positive assays results for the Piperlongumine (CID: 637858), mostly from the authors of this publication.
Out of curiosity, I performed an in silico analysis to list potential target using chemoinformatics and chemical biology tools. First I went to the PubChem and searched for chemically similar compounds (95% or more), then listed the positive bioassay results for all of the compounds. From there proceeded to the protein targets of the assays. That listed 14 potential targets.
1. TPA: Tim23p [Saccharomyces cerevisiae S288c] 222 aa protein DAA10558.1 GI:285814664
2. TPA: Tim10p [Saccharomyces cerevisiae S288c] 93 aa protein DAA06693.1 GI:285809906
3. nuclear receptor ROR-gamma [Mus musculus] 516 aa protein NP_035411.2 GI:188536040
4. LANA [Human herpesvirus 8] 1129 aa protein YP_001129431.1 GI:139472804
5. cellular tumor antigen p53 isoform a [Homo sapiens] 393 aa protein NP_000537.3 GI:120407068
6. RecName: Full=BRCA1-associated RING domain protein 1; Short=BARD-1 777 aa protein Q99728.2 GI:116241265
7. Microtubule-associated protein tau [Homo sapiens] 383 aa protein AAI14949.1 GI:92096784
8. E3 ubiquitin-protein ligase Mdm2 isoform MDM2 [Homo sapiens] 497 aa protein NP_002383.2 GI:89993689
9. protein Mdm4 isoform 1 [Homo sapiens] 490 aa protein NP_002384.2 GI:88702791
10. cytochrome P450 1A2 [Homo sapiens] 516 aa protein NP_000752.2 GI:73915100
11. Microphthalmia-associated transcription factor [Homo sapiens] 419 aa protein AAH65243.1 GI:40807040
12. aldehyde dehydrogenase 1 family, member A1 [Homo sapiens] 501 aa protein AAP35567.1 GI:30582681
13. serine/threonine kinase 33 [Homo sapiens] 514 aa protein CAC29064.1 GI:12830367
14. RecName: Full=Breast cancer type 1 susceptibility protein; AltName: Full=RING finger protein 53 1863 aa protein P38398.2 GI:728984
The interesting targets are Microtubule-associated protein tau , E3 ubiquitin-protein ligase Mdm2, Microphthalmia-associated transcription factor, BRCA1-associated RING domain protein 1 and cellular tumor antigen p53. Well, this is just a fun way to use Chemical biology and chemoinformatics for drug design.
Some SAR is listed in the US patent (US20090312373). It would be great to see more SAR of the compound analoges to study the pharmacophores.
Out of curiosity, I performed an in silico analysis to list potential target using chemoinformatics and chemical biology tools. First I went to the PubChem and searched for chemically similar compounds (95% or more), then listed the positive bioassay results for all of the compounds. From there proceeded to the protein targets of the assays. That listed 14 potential targets.
1. TPA: Tim23p [Saccharomyces cerevisiae S288c] 222 aa protein DAA10558.1 GI:285814664
2. TPA: Tim10p [Saccharomyces cerevisiae S288c] 93 aa protein DAA06693.1 GI:285809906
3. nuclear receptor ROR-gamma [Mus musculus] 516 aa protein NP_035411.2 GI:188536040
4. LANA [Human herpesvirus 8] 1129 aa protein YP_001129431.1 GI:139472804
5. cellular tumor antigen p53 isoform a [Homo sapiens] 393 aa protein NP_000537.3 GI:120407068
6. RecName: Full=BRCA1-associated RING domain protein 1; Short=BARD-1 777 aa protein Q99728.2 GI:116241265
7. Microtubule-associated protein tau [Homo sapiens] 383 aa protein AAI14949.1 GI:92096784
8. E3 ubiquitin-protein ligase Mdm2 isoform MDM2 [Homo sapiens] 497 aa protein NP_002383.2 GI:89993689
9. protein Mdm4 isoform 1 [Homo sapiens] 490 aa protein NP_002384.2 GI:88702791
10. cytochrome P450 1A2 [Homo sapiens] 516 aa protein NP_000752.2 GI:73915100
11. Microphthalmia-associated transcription factor [Homo sapiens] 419 aa protein AAH65243.1 GI:40807040
12. aldehyde dehydrogenase 1 family, member A1 [Homo sapiens] 501 aa protein AAP35567.1 GI:30582681
13. serine/threonine kinase 33 [Homo sapiens] 514 aa protein CAC29064.1 GI:12830367
14. RecName: Full=Breast cancer type 1 susceptibility protein; AltName: Full=RING finger protein 53 1863 aa protein P38398.2 GI:728984
The interesting targets are Microtubule-associated protein tau , E3 ubiquitin-protein ligase Mdm2, Microphthalmia-associated transcription factor, BRCA1-associated RING domain protein 1 and cellular tumor antigen p53. Well, this is just a fun way to use Chemical biology and chemoinformatics for drug design.
Some SAR is listed in the US patent (US20090312373). It would be great to see more SAR of the compound analoges to study the pharmacophores.
It is high time that we carry out a comprehensive systematic scientific research on the properties of traditional medicines and their chemical components. The knowledge of our forefathers should be capaitalized by us to fight the deadly diseases like cancer.
_______________________________________
Selective killing of cancer cells by a small molecule targeting the stress response to ROS
Nature Volume: 475, Pages: 231–234 (14 July 2011) DOI:doi:10.1038/nature10167
http://www.nature.com/nature/journal/v475/n7355/full/nature10167.htmlUS Patent
http://www.freepatentsonline.com/20090312373.pdf
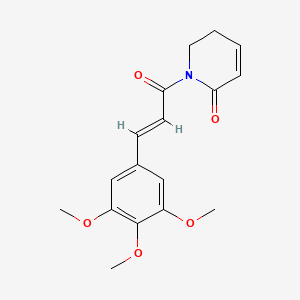
Piperlongumine
SMILES N1(C(C=CCC1)=O)C(/C=C/C2=CC(=C(OC)C(=C2)OC)OC)=O
http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=637858
http://www.indofinechemical.com/gnicart/details.aspx?catid=P-004&pid=13253
http://www.rdchemicals.com/chemicals.php?mode=details&mol_id=8163
Structural bioinformatics study: trimethoxyphenyl part in PDB
http://www.pdb.org/pdb/ligand/ligandsummary.do?hetId=SNY&sid=3P9I
Tuesday, July 19, 2011
Life science animations and videos
The Inner Life of a cell: Musical
MITOCHONDRIA
BIOVISIONS AT HARVARD UNIVERSITY
Wednesday, October 07, 2009
Nobel prize commitee recognize significance of Structure-based drug design
Structural biology has evolved considerably after the first breakthrough in understanding the molecular structure of DNA in 1950's. Nobel prize was awarded to them (Physiology/Medicine, 1962) for such an incredible discovery. This year's (2009) Nobel prize for chemistry goes to for their work on structural understanding of ribosomal machinery using X-ray crystallography. Nobel committee believe that this discovery is significant for the structure-based drug design of new antibiotics, especially since the emergence of bacterial resistance.
My thought here is "Will structure-based computational drug design methods bring some breakthrough discoveries in the next 20 years?"
Subscribe to:
Posts (Atom)